Spoiler alert: the answer is maybe! Although, my inclusion of the word “actually” betrays my bias.
Vector databases are having their day right now. Three different vector DB companies have raised money on valuations up to $700 million (paywall link). Surprisingly, their rise in popularity is not for their “original” purpose in recommendation systems, but rather as an auxillary tool for Large Language Models (LLMs). Many online examples of combining embeddings with LLMs will show you how they store the embeddings in a vector database.
While this is fine and quick for a Proof of Concept, there is always a cost to introducing new infrastructure, especially when that infrastructure is a database. I’ll talk a little about that cost in this blog post, but I’m mainly interested in whether there’s even a tangible benefit to begin with.
Why a vector database?
I think I’ll skip over the obligatory explanation of embeddings. Others have done this much better than me. So, let’s assume you know what embeddings are and that you have plans to embed some things (probably documents, images, or “entities” for a recommendation system). People typically use a vector database so that they can quickly find the most similar embeddings to a given embedding. Maybe you’ve embedded a bunch of images and want to find other dogs that look similar to a given dog. Or, you embed the text of a search query and want to find the top 10 documents that are most similar to the query. Vector databases let you do this very quickly.
Pre-meditated calculation
Vector databases are able to calculate similarity quickly because they have already pre-calculated it. Er, to be fair, they’ve approximately pre-calculated it. For $N$ entities, it takes $O(N^{2})$ calculations to calculate the similarity between every single item and every other item. If you’re Spotify and have over 100 million tracks, then this can be a pretty large calculation! (Thankfully, it’s at least embarrassingly parallel). Vector databases allow you to trade off some accuracy in exchange for speed, such that you can tractably calculate the (approximately) most similar entities to a given entity.
Do you need to pre-calculate similarity between every entity, though? I think of this like batch versus streaming for data engineering, or batch prediction vs real-time inference for ML models. One benefit of batch is that it makes real-time simple. One downside of batch is that you have to compute everything, whether or not you actually need it.
For measuring similarities, you can do this calculation in real-time. For $N$ total entities, the time complexity for calculating the top $k$ most similar entities to a given entity is $O(N)$. This comes from a couple assumptions: we’ll use cosine similarity for our similarity metric, and we’ll assume the embeddings have already been normalized. Then, for an embedding dimension $d \lt \lt N$, it’s $O(Nd)$ to calculate the similarity between a given embedding and all other $N$ embeddings. To find the top $k$ most similar entities, we have to add another $O(N + k \ log(k))$. This all nets out to roughly $O(N)$.
In numpy, the “real-time” calculation takes 3 lines of code:
# vec -> 1D numpy array of shape D
# mat -> 2D numpy array of shape N x D
# k -> number of most similar entities to find.
similarities = vec @ mat.T
partitioned_indices = np.argpartition(-similarities, kth=k)[:k]
top_k_indices = partitioned_indices[np.argsort(-similarities[partitioned_indices])]
Evidence-based claims
Depending on your size of $N$ and your latency requirements, $O(N)$ can be very reasonable! To prove my point, I put together a little benchmark. All the code for the benchmark can be found at this nn-vs-ann GitHub repo.
For my benchmark, I randomly initialize $N$ embeddings with 256 dimensions apiece. I then measure the time it takes to pick out the top 5 “nearest neighbor” (aka most similar) embeddings to a given embedding. I perform this benchmark for a range of $N$ values using two different approaches:
numpyis the “real-time” calculation that performs the full accuracy, non-precomputed nearest neighbors calculation.hnswlibuses hnswlib to pre-calculate approximate nearest neighbors.
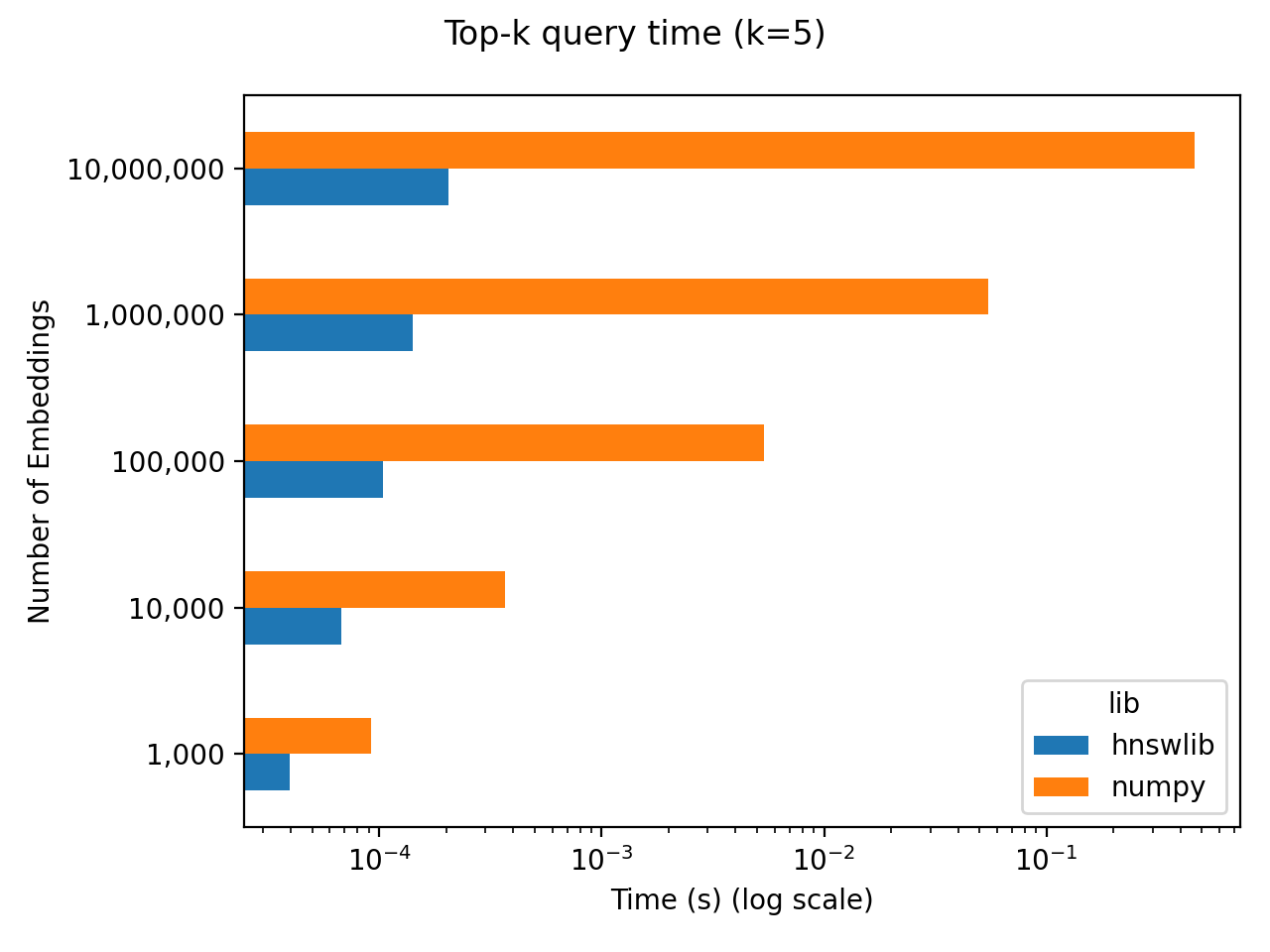
The results are shown below, with both axes on log scales. It’s hard to see when we’re dealing with log-log scales, but numpy scales linearly with $N$. The latency is roughly 50 milliseconds per million embeddings. Again, depending on your $N$ and latency requirements, 50 ms for 1 million embeddings might be perfectly fine! Additionally, you get to save yourself the complication of standing up a vector database and waiting the ~100 seconds to index those million embeddings.
But that’s not fair
Eyyyyy, you got me! I have glossed over and ignored lots of factors in this argument. Here are a couple counterarguments that may be relevant:
- The
numpyapproach requires me to hold everything in memory which is not scalable. - How do you even productionize this
numpyapproach? Pickle anarray? That sounds terrible, and then how do you update it? - What about all of the other benefits of a vector database, such as metadata filtering?
- What if I really do have a lot of embeddings?
- Shouldn’t you stop hacking things and just use the right tool for the job?
Allow me to now counter my own counterarguments:
1. The numpy approach requires me to hold everything in memory which is not scalable.
Yes, although vector databases also require holding things in memory (I think?). Also, you can acquire very high memory machines nowadays. Also also, you can memory map your embeddings if you want to trade memory for time.
2. How do you even productionize this numpy approach? Pickle an array? That sounds terrible, and then how do you update it?
I’m glad you asked! I actually did productionize this approach at a startup I worked at. Every day, I trained a contrastive learning image similarity model to learn good image representations. I wrote out the image embeddings as JSON to S3. I had an API that calculated the most similar images for an input image using the numpy method in the benchmark. That API had an async background job that would check for new embeddings on S3 every so often. When it found new embeddings, it just loaded them into memory.
3. What about all of the other benefits of a vector database, such as metadata filtering?
Yes, and this is why you should maybe just use your existing database (or even augment it!) or a tried-and-true document database like Elasticsearch rather than a vector database. Relatedly, if you need to filter by various metadata, are you already storing that metadata in your “regular” database? If so, it’s going to be annoying to have to sync that data over to a new system. I’m certainly not a fan of this.
4. What if I really do have a lot of embeddings?
Then yeah, you might just have to use a specialized vector DB (although I do think that Elasticsearch supports approximate nearest neighbors). One thing I would double check is that you can’t reduce your search to a reasonable number of embeddings prior to performing your similarity calculations. For example, if you’re searching for the most similar shirts to a given shirt, you don’t need to calculate similarities for non-shirts!
5. Shouldn’t you stop hacking things and just use the right tool for the job?
Like most things, my guess is that the right tool for the job is probably the tool you’re already using. And that tool is probably postgres, or Elasticsearch if you really need it.
Brave new world
My arguments might all be fully moot, given that we’re moving to a world of large, open-ended LLMs that need access to, like, all of Wikipedia in order to answer questions. For these applications, vector DBs surely do make sense. I’m more interested in non-open ended LLMs, though, and I do wonder how much those will require vector databases. Let’s see what happens! At the pace things are going, I can’t predict the afternoon, so I’ll just try to savor my lunch.